"RNA polymerase II (RNAPII) is responsible for the transcription of most eukaryotic protein-coding genes. Analyzing the topological distribution and quantification of RNAPII helps to understand its function in the intercellular phase. Previous studies have shown that RNAPII molecules form a network structure in the heterochromatin of differentiated Arabidopsis nuclei, rather than being organized in different "transcription factories" as observed in mammalian nuclei. ”
Measurements of immune signal intensity based on specific antibody markers in the maximum intensity projection of the image stack obtained by structured illumination microscopy (SIM) indicate an increase in the relative proportion of RNAPII in endopolyploid plant nuclei. Photoactivated localization microscopy (PALM) was applied in this study to determine the absolute number and distribution of active and inactive RNAPII molecules in differentiated Arabidopsis nuclei. The increase in the proportion of RNAPII during endopolyploidy was confirmed, as well as the fact that PALM measurements were more reliable than SIM-based measurements in terms of quantification. Single-molecule localization results show that while RNAPII molecules are globally dispersed within plant autochromatin, they also aggregate over smaller distances as described by mammalian transcription factories.
《Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei》
01Research Results (excerpt)
1. Combine SIM and PALM to super-resolution imaging of RNAPII in Arabidopsis nuclei
Most eukaryotic genes are transcribed by RNAPII, and the amount of active RNAPII in the nucleus reflects the degree of transcription. Colocalization of transcripts and RNAPII was demonstrated by electron and optical microscopy. Depending on its position on the gene and transcription stage, RNAPII is phosphorylated to varying degrees, while inactive RNAPII is mainly unphosphorylated. In mammals, RNAPII is thought to be organized in different so-called "transcription factories", and studies have found that the high variability in the diameter of the transcription factory and the number of RNAPII molecules in it is caused not only by the different cell types analyzed, but also by the different imaging methods applied, while super-resolution microscopy techniques, such as structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM), locate individual molecules with high precision beyond the diffraction limit of light, Molecules in cells can be counted more reliably.
The nucleus is labeled with specific antibodies against active and inactive RNAPII, and SIM is applied in conjunction with 3D-PALM. After determining the distribution of RNAPII enzymes in the SIM image, the network structure becomes visible. The number and localization of individual scintillation molecules is recorded by 3D-PALM. When the molecules are rendered to the acquired localization accuracy, active and inactive RNAPII molecules are evenly distributed throughout the nucleus, except for nucleoli that lack RNAPII. The combination of SIM and PALM and simultaneous labeling of different RNAPII molecules with the fluorophores Alexa488 and Cy5 in the same nucleus allow individual molecules to localize within the network structure. No colocalization of active and inactive RNAPII occurred, indicating that the two molecules occupy different regions within autochromatin.
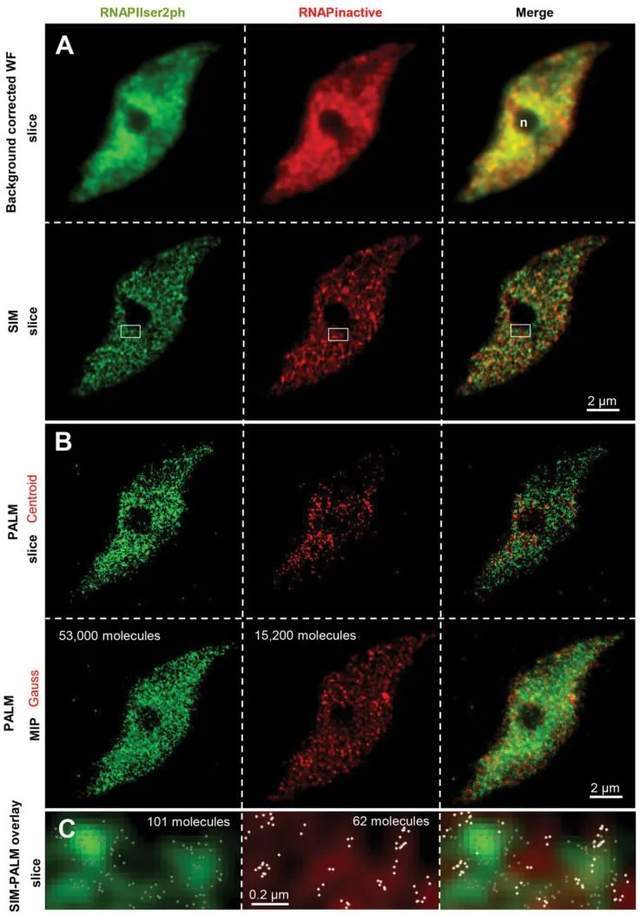 Figure 1. Distribution of RNAPII molecules in Arabidopsis leaf nuclei
Figure 1. Distribution of RNAPII molecules in Arabidopsis leaf nuclei
2. Use 3D-PALM to analyze the distribution of RNAPII molecules
Super-resolution microscopy can also assess the third dimension of biological specimens, and the number of RNAPII molecules in the nuclei of differentiated endopolyploid plants was determined for the first time in this study by applying 3D-PALM. Due to the presence of more molecules, active RNAPII shows a higher global density than inactive. Both RNAPIIs exhibit similar aggregation behavior and regularly present high accumulation sites. For active and inactive RNAPII, the distance between individual RNAPII molecules within small clusters is between 20 nm and 40 nm. For active and inactive RNAPII, there are regions of further accumulation between small clusters. Thus, in addition to the global dispersion of two RNAPII molecules within the autochromatin, RNAPII molecules may also aggregate at two different levels. Despite an average single-molecule distance of 27 nm, active RNAPII molecules are more densely clustered in small- and large-distance clusters than inactive molecules. Size clusters correspond to the size of known animal transcription factories.
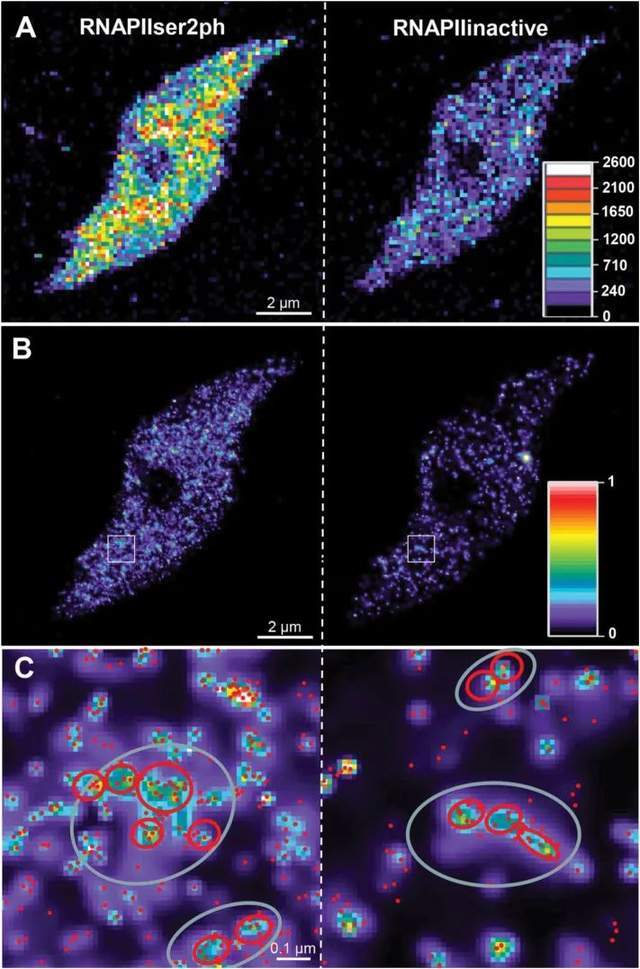 Figure 2. Distribution of inactive and active RNAPII molecules in 8C Arabidopsis nuclei recorded by 3D-PALM
Figure 2. Distribution of inactive and active RNAPII molecules in 8C Arabidopsis nuclei recorded by 3D-PALM
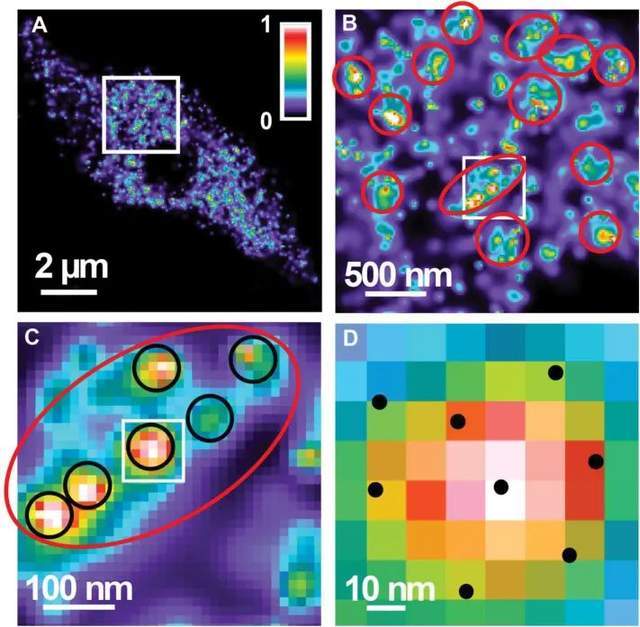 Figure 3. Distribution and aggregation of active RNAPII (Ser2ph).
Figure 3. Distribution and aggregation of active RNAPII (Ser2ph).
3. The relationship between the number of RNAPII molecules and endopolyploidy
Using 3D-PALM, the researchers also estimated the copy number of active and inactive RNAPIIs throughout the nucleus. The average number of active RNAPII (Ser2ph) increases in differentiated 2C and endopolyploid 4C-16C leaf nuclei, ranging between ~13000 in 2C and ~58000 in 16C. However, in the case of precise enzymatic replication during intranuclear replication, the number of molecules does not increase proportionally as theoretically expected.
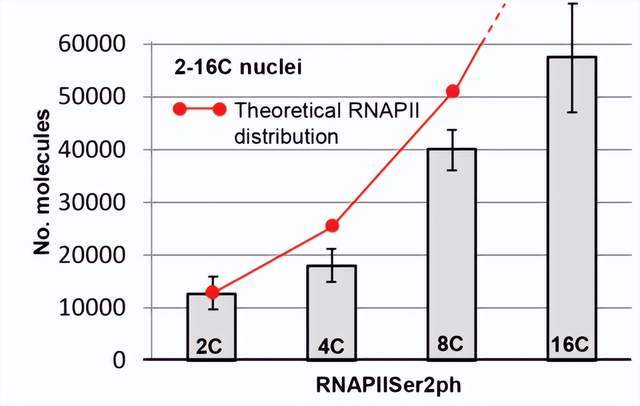 Figure 4. Average number of active RNAPII (Ser2ph) in 2C–16C nuclei
Figure 4. Average number of active RNAPII (Ser2ph) in 2C–16C nuclei
4. A tissue model of endopolyploid plant interphase nucleus
Previous findings have shown that chromosomal structure maintenance agglutinin (SMC) and agglutinin complex subunits SMC3 and CAP-D3 show RNAPII-like reticular distributions within differentiated endopolyploid Arabidopsis nuclear autochromatin, respectively. Based on this, the researchers proposed a model (Figure 5): two different adjacent chromosomal arm regions (blue and white) with a high concentration of heterochromatin. The autochromatin of sister chromatids consists of chromatin fragments of about 50 kb, which are further aggregated. Sister chromatid fragments can be cohesive or separated. Chromatin fibers may originate where RNAPII molecules aggregate and are activated in potential transcription factories. Most of the inactive RNAPII molecules are also aggregated and distributed in autochromatin. The SMC3 and CAP-D3 subunits are evenly distributed and may be responsible for maintaining flexible autochromatin tissue.
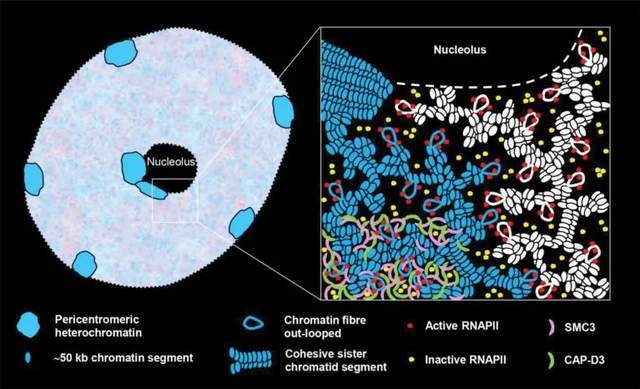 Figure 5. Model of the arrangement of RNAPII and SMC protein complex subunits (SMC3 and CAP-D3) within interphase plant nuclear autochromatin
Figure 5. Model of the arrangement of RNAPII and SMC protein complex subunits (SMC3 and CAP-D3) within interphase plant nuclear autochromatin
02 Research Summary
1. A network structure formed by different RNAPIIs, containing some major globally dispersed single molecules that may aggregate into the size of known animal "transcription factories";
2. Quantitative RNAPII by 3D-PALM is more reliable than signal intensity measurement of the maximum intensity projection generated by the SIM image stack;
3.3D-PALM measurements confirm that the number of RNAPII molecules increases with the degree of endopolyploidy.
References
1.Schubert, Veit, and Klaus Weisshart. "Abundance and distribution of RNA polymerase II in Arabidopsis interphase nuclei." Journal of experimental botany 66.6 (2015): 1687-1698.
In this study, researchers mainly use 3D-PALM super-resolution microscope to study RNAPII, 3D-PALM and 3D-STORM principle is similar, at present, in China, random optical reconstruction microscope STORM has been successfully commercialized, there are experts and teachers who need STORM imaging technology for experimental research, please fill in the questionnaire at the end of the text, you can make an appointment to get iSTORM super-resolution microscopy imaging system test shooting service~

iSTORM, the ultra-high-resolution microscopic imaging system released by Lixian Intelligence, has successfully achieved a breakthrough in the diffraction limit of optical microscopy, making it possible to engage in the research of single-molecule localization and counting of biological macromolecules, subcellular and macromolecular complex structure analysis, and biodynamics of biological macromolecules on the resolution scale of 20 nm, thus bringing major breakthroughs to life sciences, medicine and other fields.
About us
Ningbo Lixian Intelligent Technology Co., Ltd. (INVIEW) is a scientific and technological enterprise specializing in ultra-high resolution microscopy technology and product research and development, relying on the automatic control, new generation information technology of Fudan University and the biology, optics, image processing and other technologies of the Hong Kong University of Science and Technology, with optics, biology, automatic control, machinery, information technology and other interdisciplinary technical teams, the 2014 Nobel Prize in Chemistry technology industrialization, launched the ultra-high resolution microscopy imaging system iSTORM, A series of products such as the cell intelligent monitoring assistant Celeston Micro help people observe the microscopic world from an unprecedented perspective, break through the limit, and see things like never before.


