Imaging of intracellular pathogens within host cells is complicated by the low resolution and sensitivity of fluorescence microscopy and the lack of ultrastructural information to observe pathogens. Here, the authors propose a novel way to observe how these pathogens behave during infection, thus avoiding these problems: by bioorthogonal labeling of the intracellular pathogen Salmonella typhimurium, and by using these bioorthogonal sets to introduce fluorophores compatible with the random optical reconstruction microscope STORM and place them in an associated photoelectron microscopy (CLEM) workflow, pathogens can be imaged in their host cell environment, Typhimurium with a resolution of 20 nm. Therefore, the STORM-CLEM method provides a new way to understand the infection process of these pathogens.

Research Introduction (Excerpt)
The rise of antibiotic resistance is one of the major threats to global health. In this regard, a class of pathogens proved to be particularly troublesome, such as intracellular bacteria. They hinder immunoassays by residing and replicating in host cell phagosomes. By secreting factors that manipulate phagosome maturation, they ensure their survival within cells, although mammalian hosts deploy a large number of defense mechanisms, so understanding bacterial-host interactions at the cellular and molecular levels is extremely important. Bioorthogonal chemistry has proven to be a major breakthrough technique for studying these host-pathogen interactions. However, the labeling methods we are currently trying either require mutant tRNA/tRNA synthetase pairs specific for bioorthogonal methionine, phenylalanine, or n-leucine analogues for the introduction of pathogens into pathogens to achieve incorporation of desired groups, or have low sensitivity. This limits them to using only those bacterial strains for which the technology can be used.
In the past few years, super-resolution imaging technology has boomed, achieving resolution at the nanoscale . Building on previous research, the authors have added super-resolution microscopy that will help overcome pathogen-related obstacles: the stochastic optical resolution microscope STORM has experimental sensitivity and improved resolution that brings the resolution of the fluorescence signal closer to that of TEM and improves detection sensitivity, allowing imaging of genetically unmodified pathogenic bacteria by CLE M.
Research results (excerpt)
 Fig. 1 Flow cytometry analysis of Hp-S. typhoid markers and their persistence in vitro and intracellular settings.
Fig. 1 Flow cytometry analysis of Hp-S. typhoid markers and their persistence in vitro and intracellular settings.
Typhoid fever is incubated with Met (4 mm) or Hpg (0.04-4 mm), then immobilized and ccHc with Alexa-647-azide. (B) Incubate Salmonella typhimurium with 0.4 mmHpg for 30 min by incubation and measure ccHc signal at the specified time. (C) showing DsRed expression as a measure of the total number of bacteria (D) showing that the ccHc signal persists for 0-3 h.
We next used the STORM-CLEM method to detect Hpg-Salmonella within BM-DC (Figure 2).
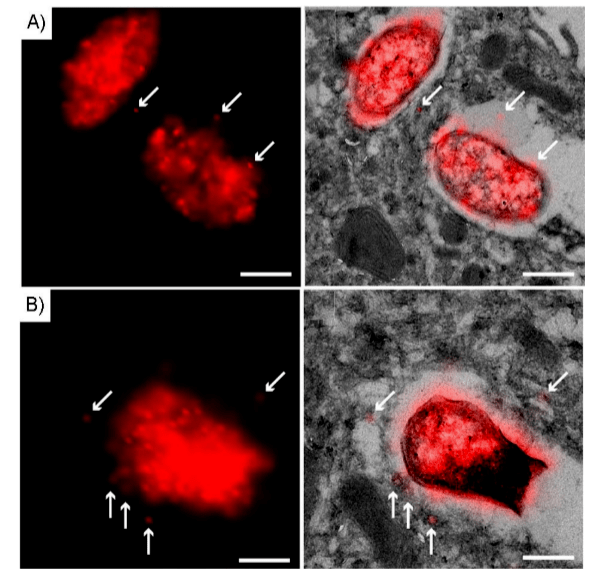 Figure 2 Super-resolution N-STORM-CLEM image of BM-DC 75 nm cryosection processed with Hpg-S.
Figure 2 Super-resolution N-STORM-CLEM image of BM-DC 75 nm cryosection processed with Hpg-S.
Typhoid incubates BM-DC with Hpg-S. Wash with phosphate buffer to remove unbound/uninternalized S. Typhoid to fix cells and perform Tokuya sample preparation and cryosectioning into 75 nm sections. Under ccHc conditions (red), treat sections with Alexa-647-azide. Left: Examples of Hpg-S super-resolution N-STORM imaging A)1 and B)2. Typhoid right: Super resolution CLEM image from left panel. Arrows indicate phagocytosis signals. Scale bar: 250 nm.
The results showed that the accuracy and detection sensitivity of fluorescent labeled Hpg-S were better than those of the control group. The infection rate of Salmonella typhimurium in STORM images was significantly higher than that in low-resolution confocal images. Subsequent observation of the intracellular environment by electron microscopy showed that STORM did not disrupt the ultrastructure of phagocytes. The membrane was found to be intact and no structural changes were observed between the sample regions imaged by STORM relative to those areas that were not analyzed by STORM. Organelles are easily recognizable based on their unique morphological appearance. The correlation of the STORM images suggests that the bioorthogonal signal is mainly on small structures with a diameter of about 10-20 nm around Salmonella or parasitic vacuoles.
This is the first example of super-resolution stochastic optical reconstruction microscopy (STORM)-correlated photoelectron microscopy (CLEM) using bioorthogonal sets and its application in the study of host-pathogen interactions, and can therefore be used to observe non-transgenic pathogens in spatially detailed host environments. Compared to the authors' method and previously reported methods, the sensitivity of STORM even allows the detection of extracellular protein products. This opens up the possibility of imaging pathogens for which previous bioorthogonal non-normative amino acid labeling (BONCAT) methods were ineffective. The combination of STORMCLEM with BONCAT, tRNA/tRNA synthetase mutants, or bioorthogonal cell wall markers will be valuable for studying the in vivo life cycle of these pathogens. Applying this technique to other areas where bioorthogonal chemistry has already been transformed will also add an additional ultrastructural dimension to these fields.
References
[1] G. Mitchell, R. R. Isberg, Cell Host Microbe 2017, 22,166–175.
[2] S. Pandey, T. Kawai, S. Akira, Cold Spring Harbor Perspect. Biol. 2015, 7,a016246.
[3] a) M. S. Siegrist, S. Whiteside, J. C. Jewett, A. Aditham,F.Cava, C. R. Ber-tozzi, ACS Chem. Biol. 2013, 8,500–505;b)P.Shieh,M.S.Siegrist, A. J.Cullen, C. R. Bertozzi, Proc. Natl. Acad. Sci. USA 2014, 111,5456–5461;c) G. W. Liechti, E. Kuru, E. Hall, A. Kalinda,Y.V.Brun, M. VanNieuwenhze,A. T. Maurelli, Nature 2014, 506,507–510.
[4] a) B. M. Swarts, C. M. Holsclaw, J. C. Jewett, M. Alber,D.M.Fox, M. S.Siegrist, J. A. Leary,R.Kalscheuer,C.R.Bertozzi, J. Am. Chem. Soc. 2012,134,16123–16126; b) K. M. Backus,H.I.Boshoff, C. S. Barry,O.Boutur-eira, M. K. Patel, F. D’Hooge, S. S. Lee, L. E. Via, K. Tahlan, C. E. Barry,B.G.Davis, Nat. Chem.Biol. 2011, 7,228–235;c)F.P.Rodriguez-Rivera, X.Zhou, J. A. Theriot, C. R. Bertozzi, J. Am. Chem. Soc. 2017, 139,3488–3495.
[5] D. C. Dieterich, A. J. Link, J. Graumann, D. A. Tirrell, E. M. Schuman, Proc.Natl. Acad. Sci. USA 2006, 103,9482–9487.
[6] A. Mahdavi,J.Szychowski, J. T. Ngo, M. J. Sweredoski, R. L. Graham, S.Hess, O. Schneewind,S.K.Mazmanian, D. A. Tirrell, Proc. Natl. Acad. Sci.USA 2014, 111,433–438.
[7] a) M. Grammel, P. D. Dossa, E. Taylor-Salmon,H.C.Hang, Chem.Commun. 2012, 48,1473–1474;b)M.Grammel, M. M. Zhang, H. C.Hang, Angew.Chem. Int. Ed. 2010, 49,5970–5974; Angew.Chem. 2010,122,6106–6110; c) S. Lin, Z. Zhang, H. Xu, L. Li, S. Chen, J. Li, Z. Hao,P. R. Chen, J. Am. Chem. Soc. 2011, 133,20581–20587.
[8] J. T. Ngo, J. A. Champion, A. Mahdavi, I. C. Tanrikulu, K. E. Beatty, R. E.Connor,T.H.Yoo,D.C.Dieterich,E.M.Schuman,D.A. Tirrell, Nat.Chem.Biol. 2009, 5,715–717.
[9] A. G. Chande, Z. Siddiqui,M.K.Midha, V. Sirohi, S. Ravichandran, K. V. S.Rao, Sci. Rep. 2015, 5,13430.
[10]R.Hatzenpichler,S.A.Connon,D.Goudeau,R. R. Malmstrom,T.Woyke,V. J. Orphan, Proc. Natl. Acad. Sci. USA 2016, 113,E4069–E4078.
At present, in China, the random optical reconstruction microscope STORM has been successfully commercialized, and experts and teachers who need STORM imaging technology for experimental research, please fill in the questionnaire at the end of the article to make an appointment to obtain the iSTORM super-resolution microscopy imaging system test shooting service~

iSTORM, the ultra-high-resolution microscopic imaging system released by Lixian Intelligence, has successfully achieved a breakthrough in the diffraction limit of optical microscopy, making it possible to engage in the research of single-molecule localization and counting of biological macromolecules, subcellular and macromolecular complex structure analysis, and biodynamics of biological macromolecules on the resolution scale of 20 nm, thus bringing major breakthroughs to life sciences, medicine and other fields.
About us
Ningbo Lixian Intelligent Technology Co., Ltd. (INVIEW) is a scientific and technological enterprise specializing in ultra-high resolution microscopy technology and product research and development, relying on the automatic control, new generation information technology of Fudan University and the biology, optics, image processing and other technologies of the Hong Kong University of Science and Technology, with optics, biology, automatic control, machinery, information technology and other interdisciplinary technical teams, the 2014 Nobel Prize in Chemistry technology industrialization, launched the ultra-high resolution microscopy imaging system iSTORM, A series of products such as the cell intelligent monitoring assistant Celeston Micro help people observe the microscopic world from an unprecedented perspective, break through the limit, and see things like never before.


